 v4.3
enviMass
v4.3
enviMass
 v4.3
enviMass
v4.3
enviMass
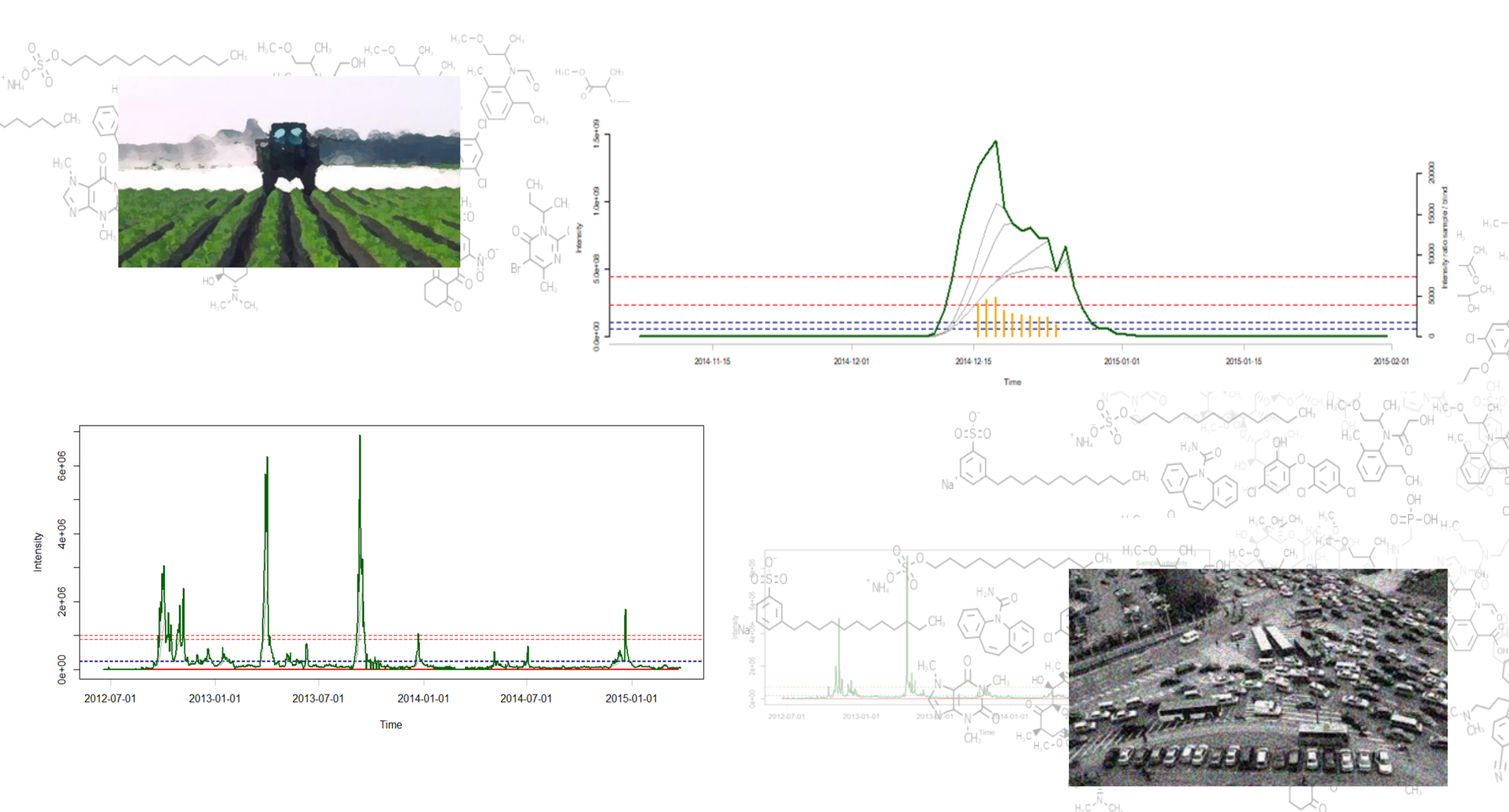
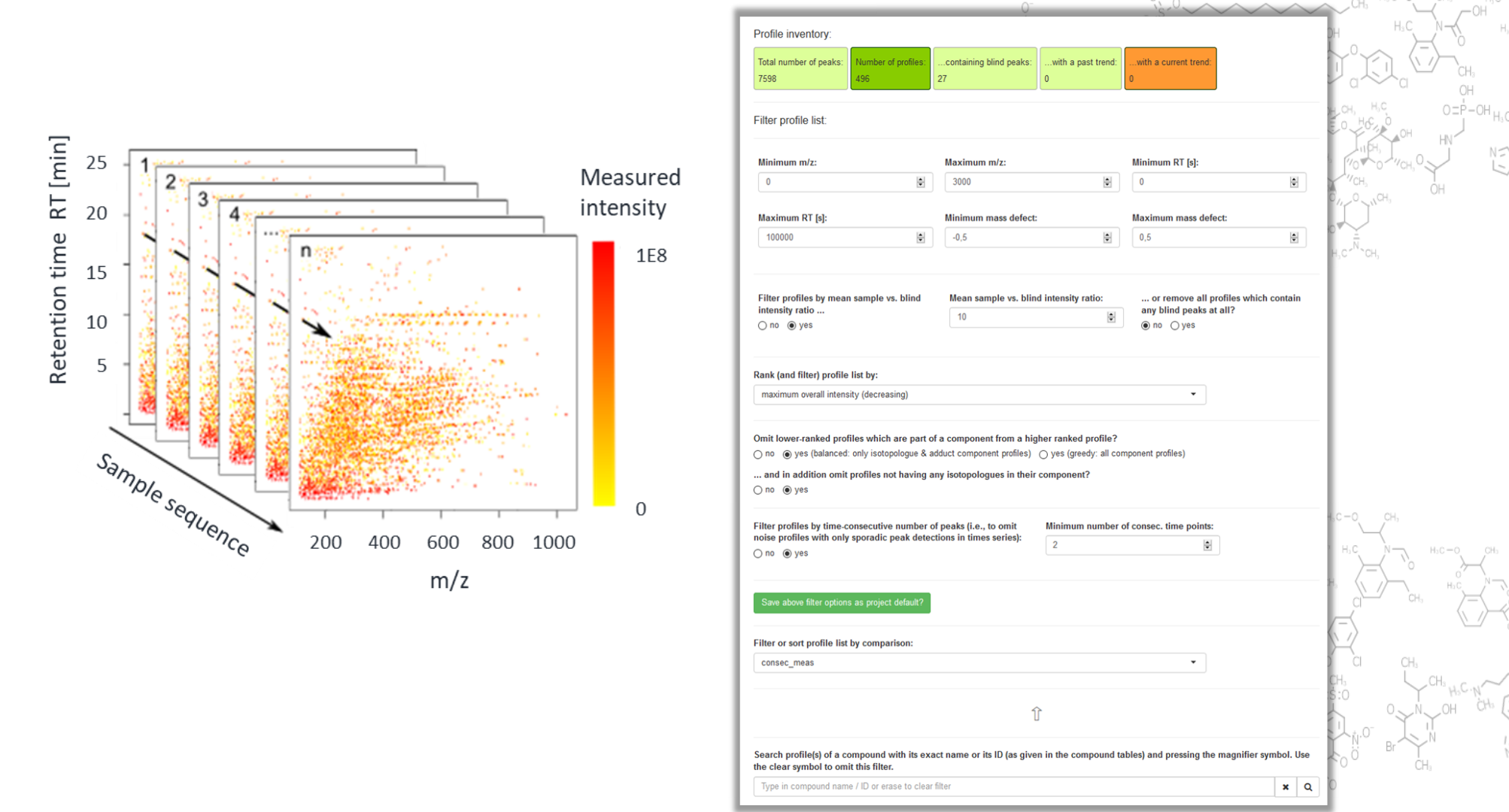
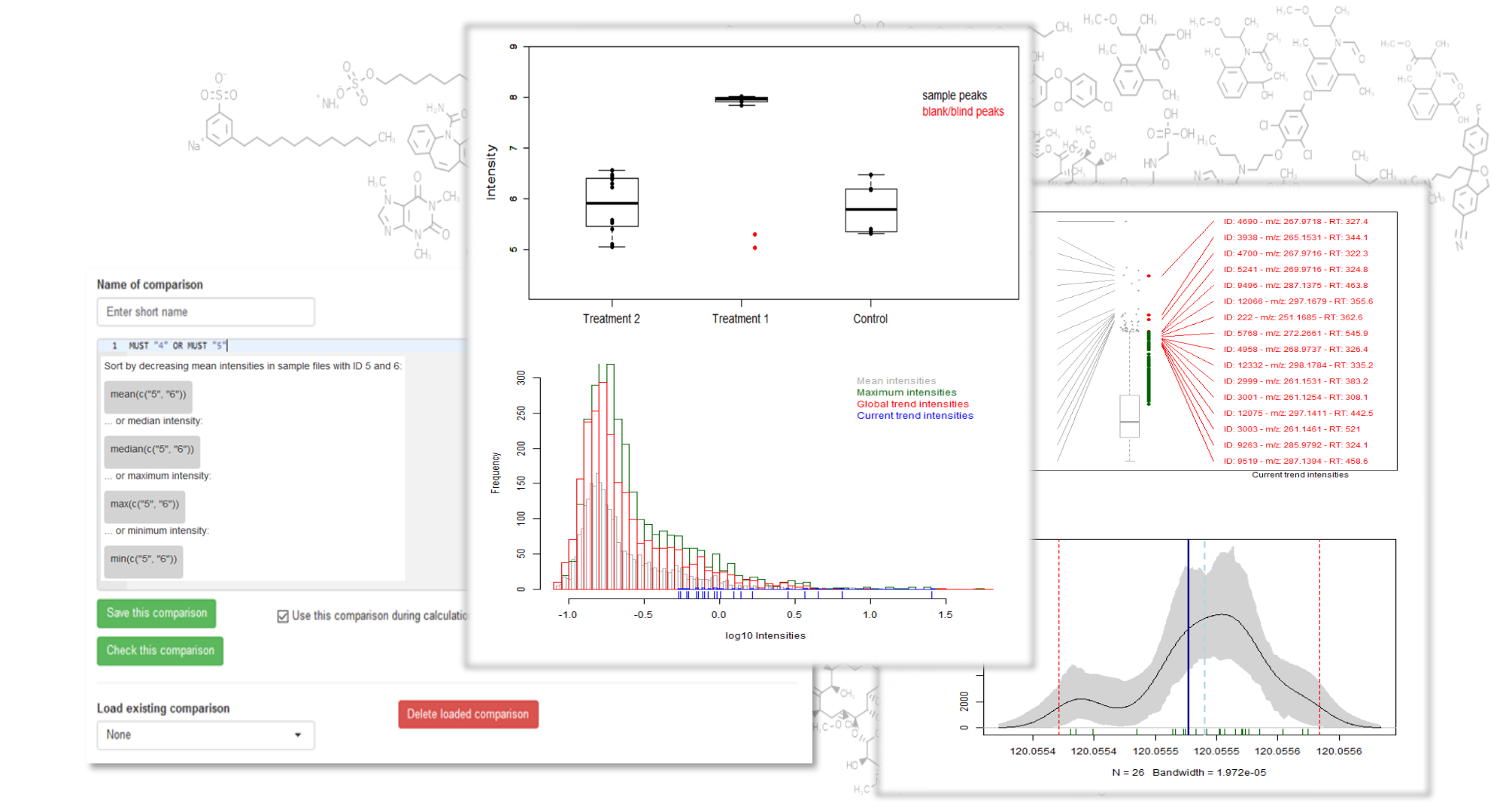
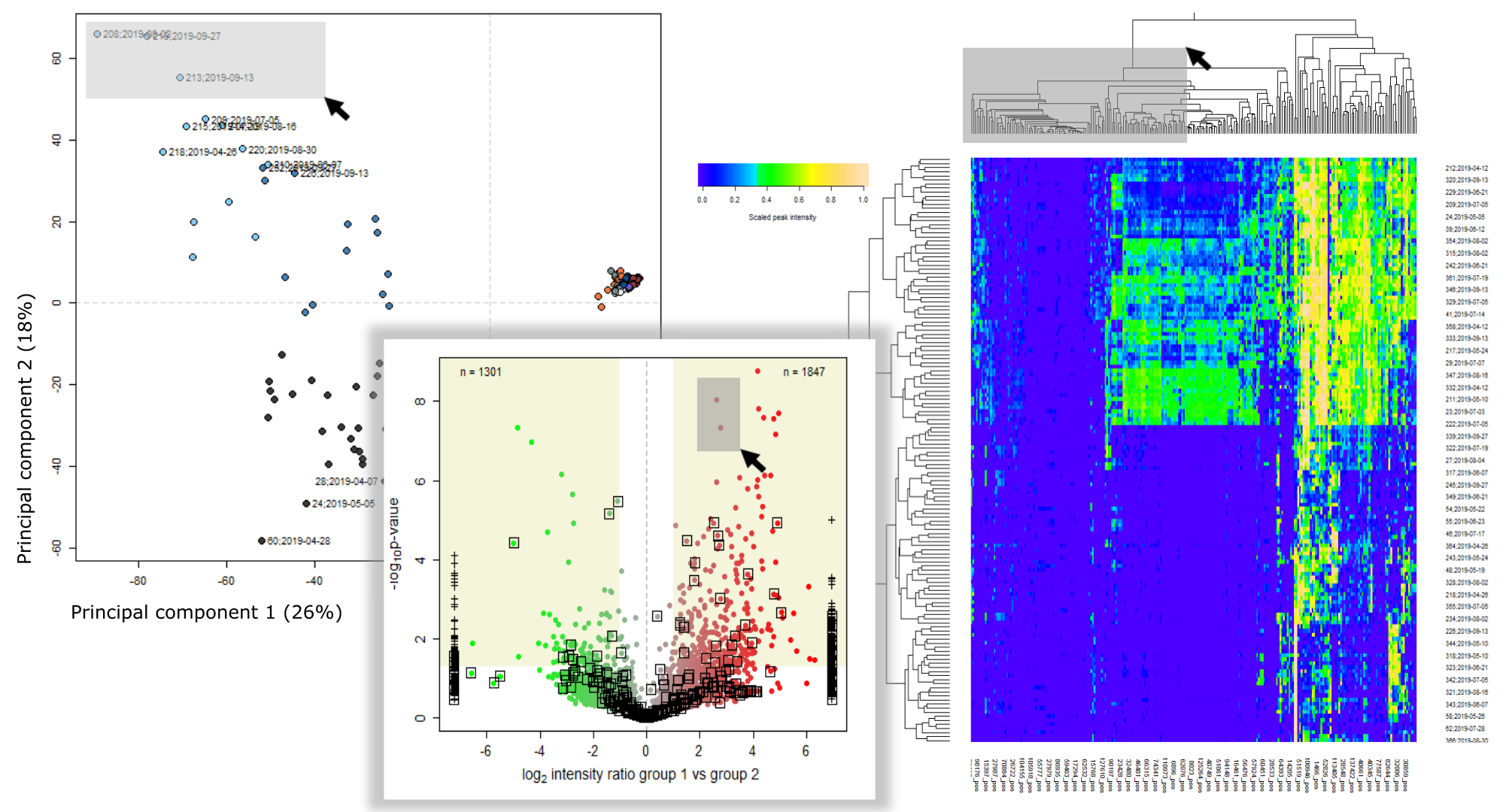
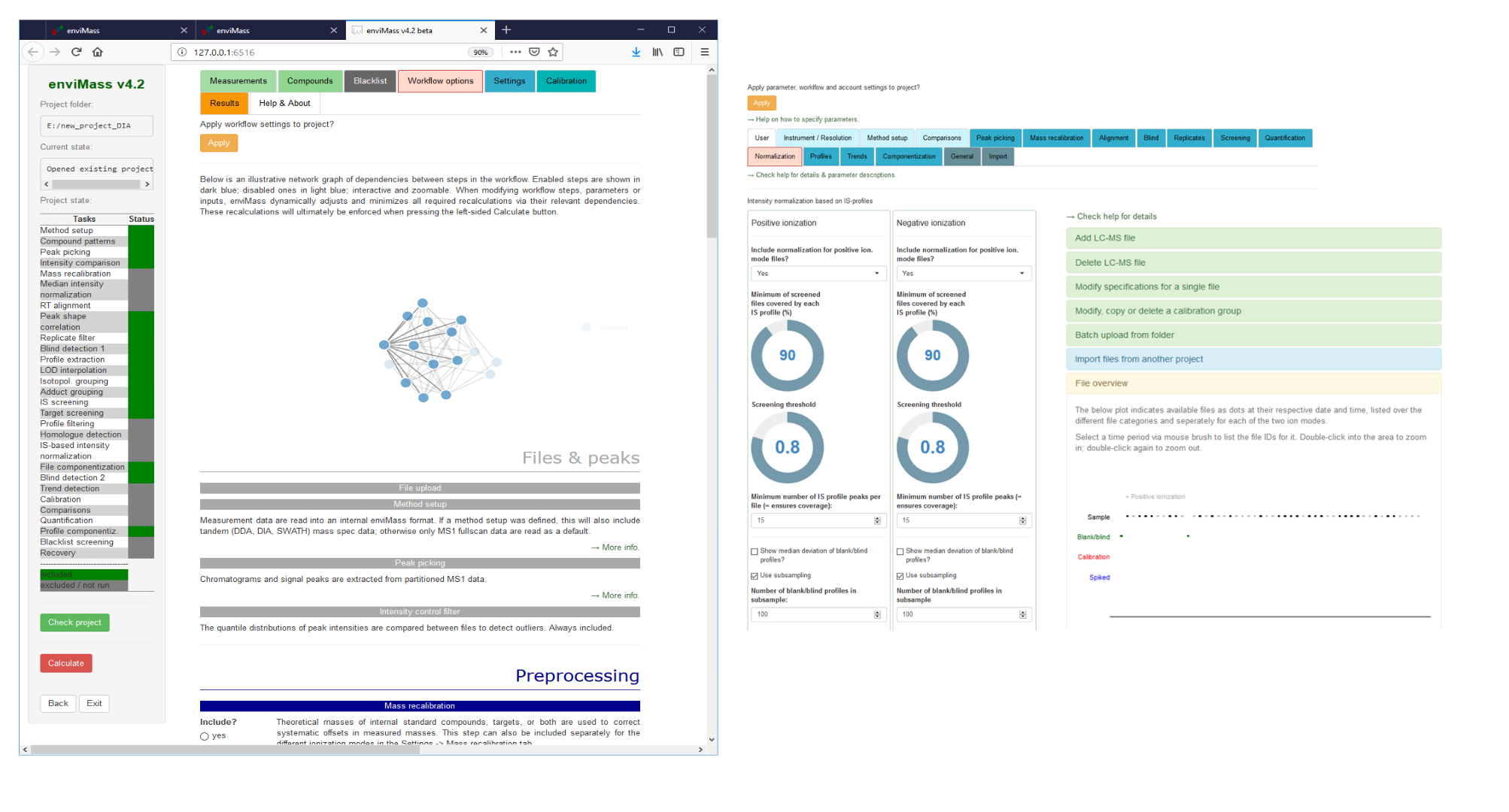
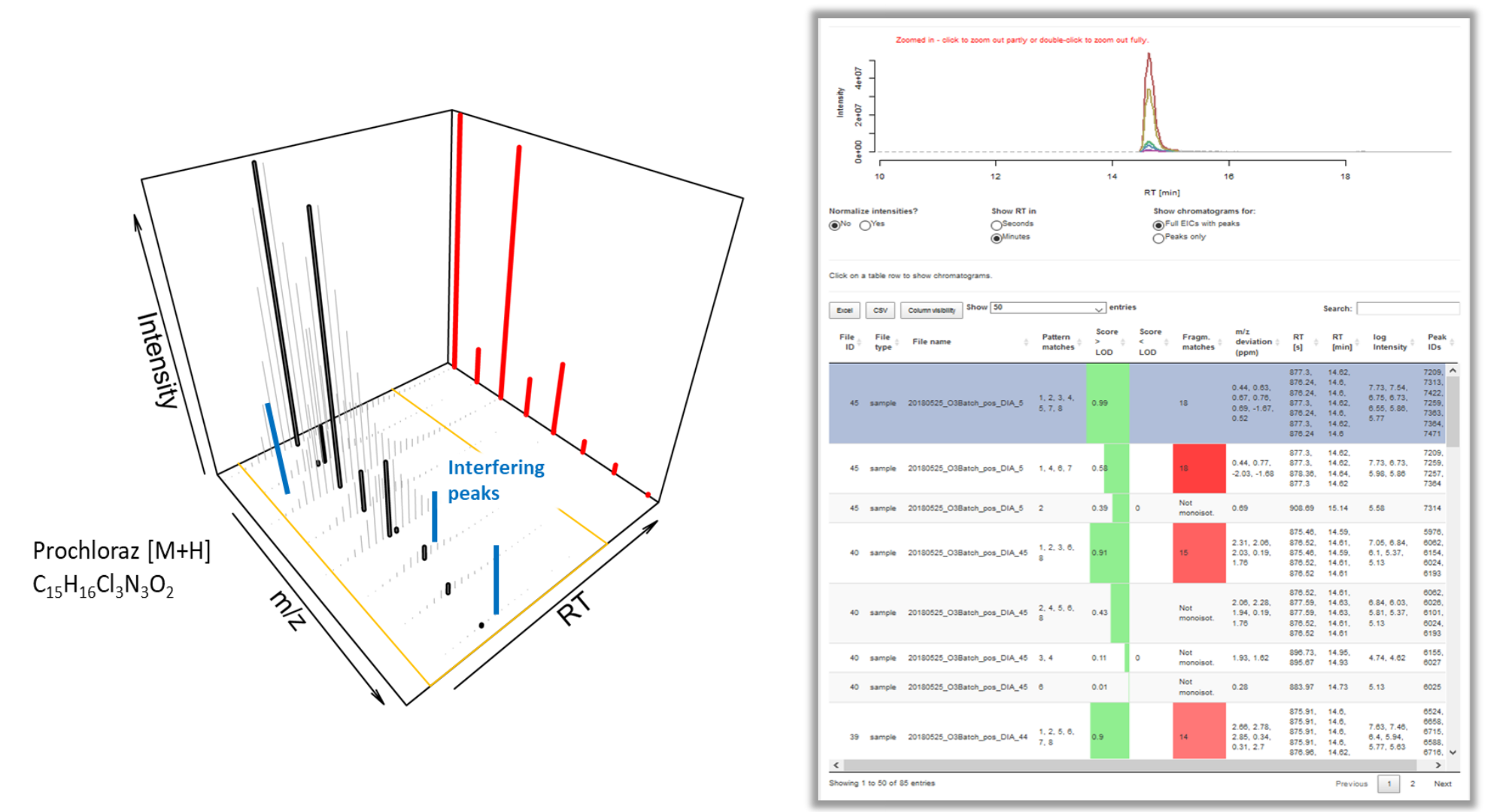
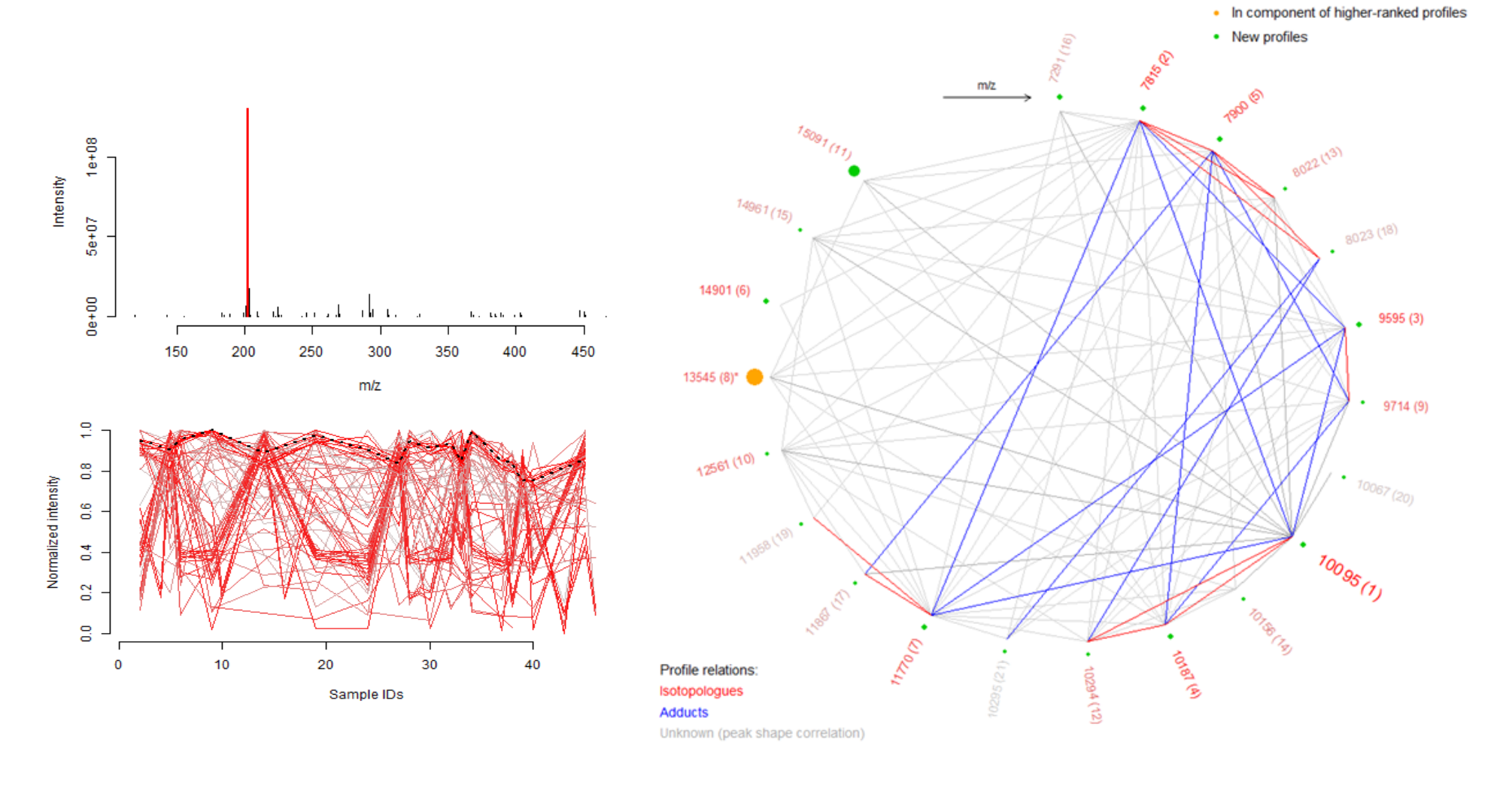
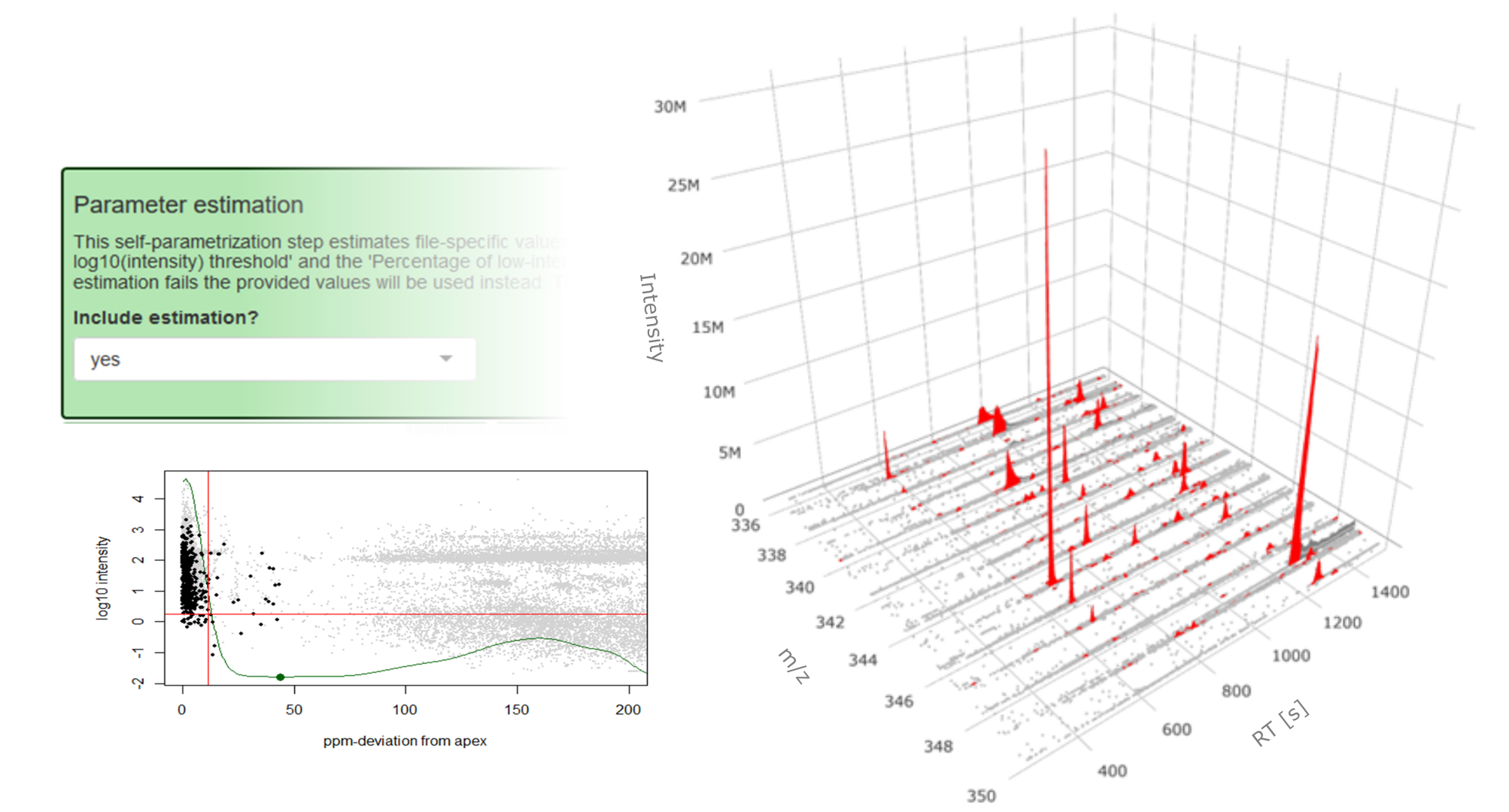
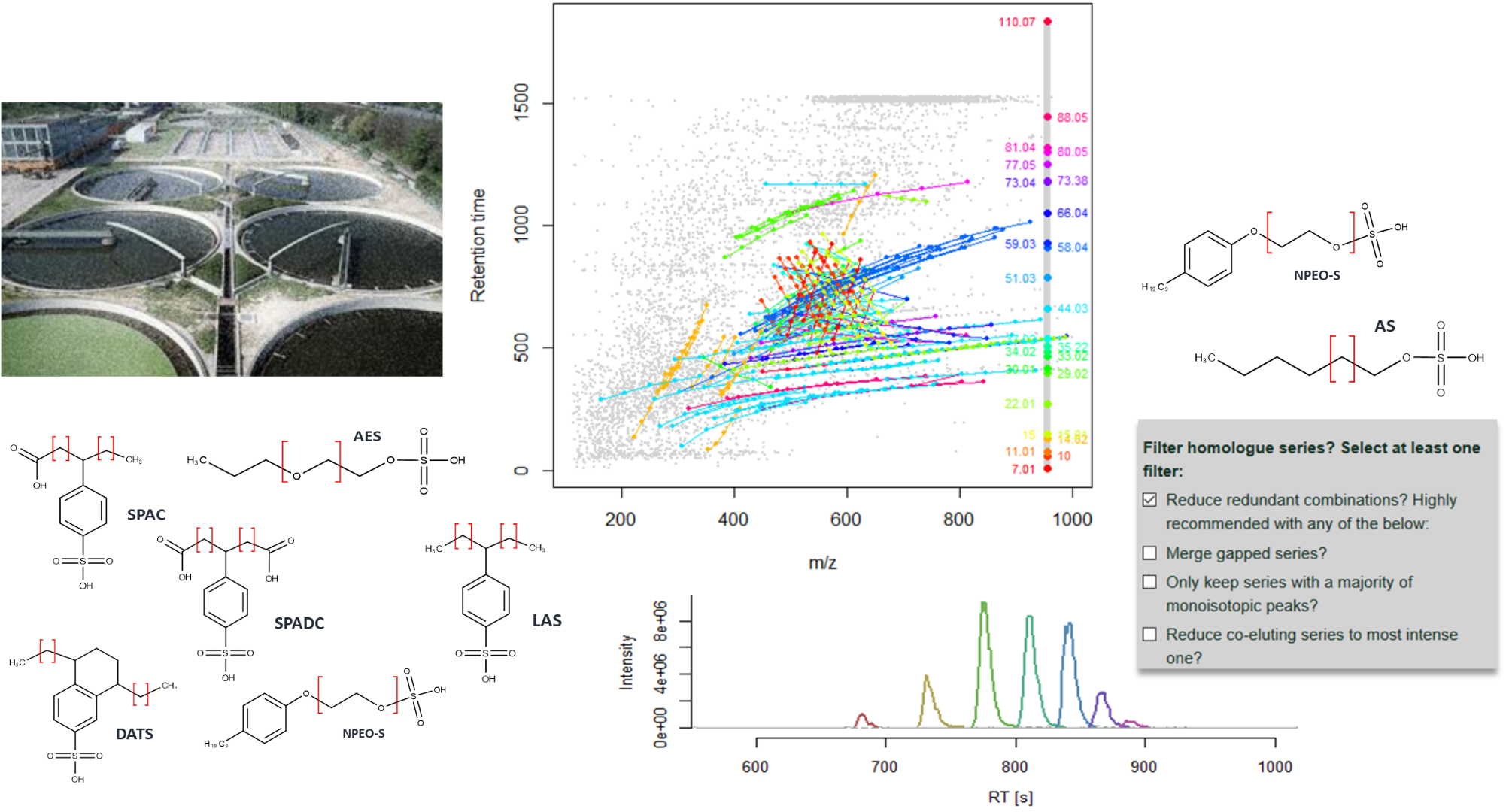
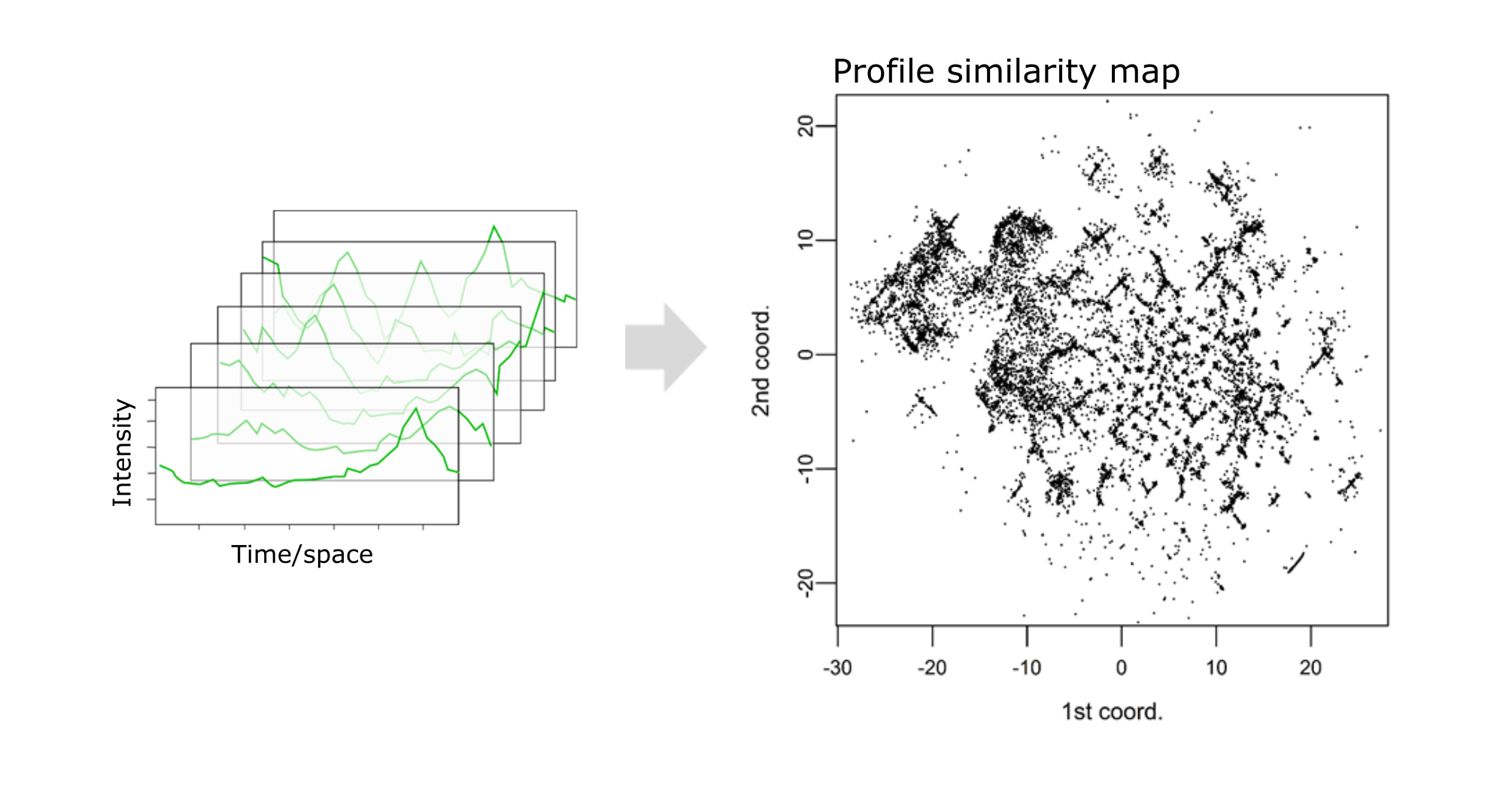
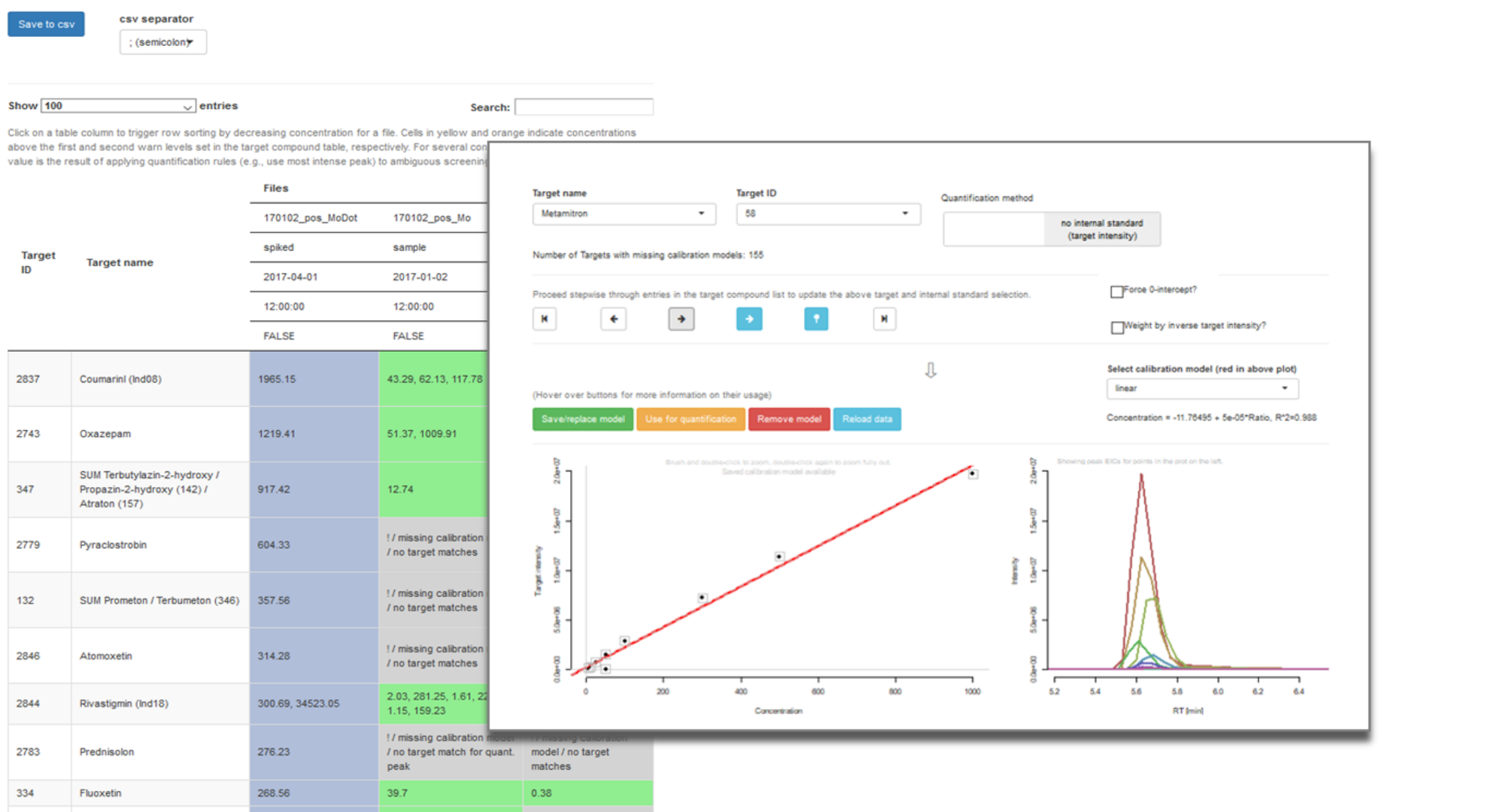


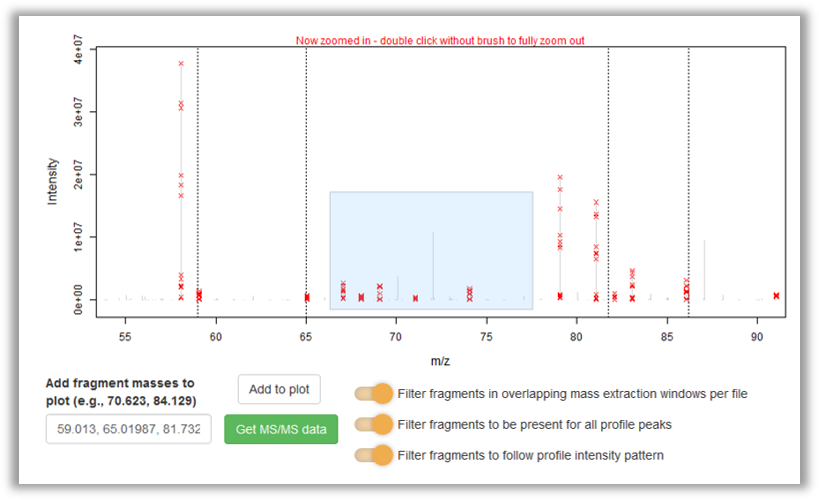
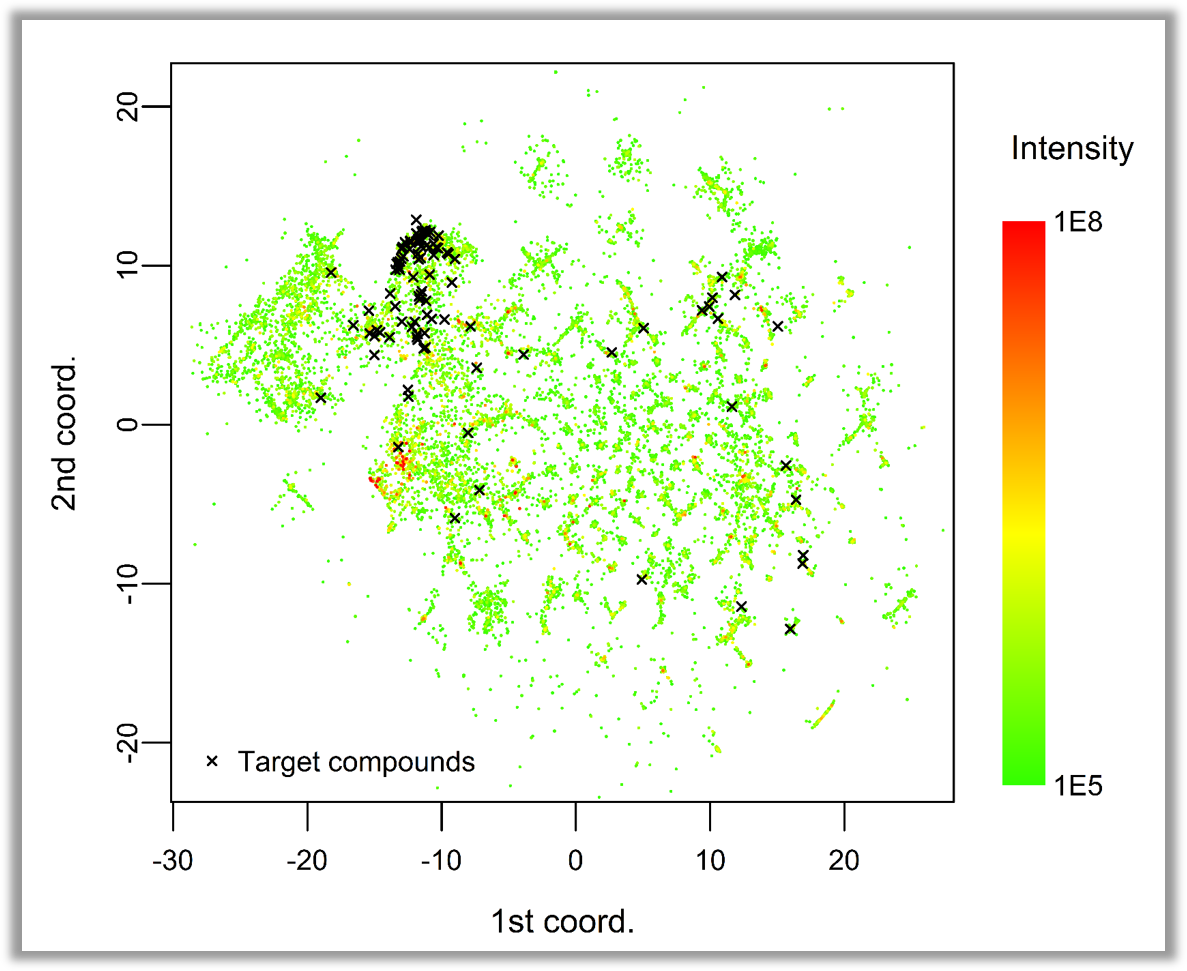
Unveiling industrial emissions in a large European river: Insights from data mining of high-frequency measurements, T. Chonova, S. Ruppe, I. Langlois, D. Griesshaber, M. Loos, M. Honti, K. Fenner, H. Singer, Water Research Volume 268, Part B, 1 January 2025, 122745
Organische Spurenstoffe – Emerging Pollutants. Untersuchung von Deponiesickerwasser, AWEL – Amt für Abfall, Wasser, Energie und Luft, Kanton Zürich, 2023
Retrospective non-target analysis to support regulatory water monitoring: from masses of interest to recommendations via in silico workflows, A. Lai, R. Singh, L. Kovalova, O. Jaeggi, T. Kondic, E. Schymanski, Environmental Sciences Europe, 2021, 33, Article number 43.
Characterization of water-soluble synthetic polymeric substances in wastewater using LC-HRMS/MS, T. Mairinger, M. Loos, J. Hollender, Water Research 190, 2021.
Assessing emissions from pharmaceutical manufacturing based on temporal high-resolution mass spectrometry data, S. Anliker, M. Loos, R. Comte, M. Ruff, K. Fenner, H. Singer, Environ. Sci. Technol., 2020, 54, 7, 4110–4120.
Application of immobilized TiO2 on PVDF dual layer hollow fibre membrane to improve the photocatalytic removal of pharmaceuticals in different water matrices, L. Paredes, S. Murgolo, H. Dzinun, M. Othmanc, A. Ismail, M. Carballa, G. Mascolo, Applied Catalysis B: Environmental 240, 2019, 9–18.
Biotransformation of antibiotics: Exploring the activity of extracellular and intracellular enzymes derived from wastewater microbial communities, M. Zumstein, D. Helbling, Water Research 155, 2019, 115-123.
Fall creek monitoring station: highly resolved temporal sampling to prioritize the identification of nontarget micropollutants in a small stream, C. Carpenter, L. Wong, C. Johnson, D. Helbling, Environ. Sci. Technol. 2019, 53, 77−87.
Photocatalytic degradation of diclofenac by hydroxyapatite–TiO2 composite material: identification of transformation products and assessment of toxicity, S. Murgolo, I. Moreira, C. Piccirillo, P. Castro, G. Ventrella, C. Cocozza, G. Mascolo, Materials 2018, 11, 1779.
Surface water and groundwater analysis using aryl hydrocarbon and endocrine receptor biological assays and liquid chromatography-high resolution mass spectrometry in Susquehanna County, PA, M. Bamberger, M. Nell, A. Ahmed, R. Santoro, A. Ingraffea, R. Kennedy, S. Nagel, D. Helbling, R. Oswald Environ Sci Process Impacts. 2019 May 16.
Comparison of different types of landfill leachate treatments by employment of nontarget screening to identify residual refractory organics and principal component analysis, C. Pastore, E. Barca, G. Del Moro, C. Di Iaconi, M. Loos, H. Singer, G. Mascolo, Science of the Total Environment 635, 2018, 984–994.
Comprehensive micropollutant screening using LC-HRMS/MS at three riverbank filtration sites to assess natural attenuation and potential implications for human health, J. Hollender, J. Rothardt, D. Radny, M. Loos, J. Epting, P. Huggenberger, P. Borer, H. Singer, Water Research X 1, 2018, 100007.
Detective work on the Rhine River in Basel – finding pollutants and polluters, S. Ruppe, D. Griesshaber, I. Langlois, H. Singer, J. Mazacek CHIMIA 2018, 72, No. 7/8.
Nitrate determines growth and protease inhibitor content of the cyanobacterium Microcystis aeruginosa, C. Burberg, M. Ilić, T. Petzoldt, E. Elert, Journal of Applied Phycology, 2018, https://doi.org/10.1007/s10811-018-1674-0.
Non-target screening to trace ozonation transformation products in a wastewater treatment train including different post-treatments, J. Schollee, M. Bourgin, U. Gunten, C. McArdell, J. Hollender, Water Research 142, 2018, 267-278.
Exploring micropollutant biotransformation in three freshwater phytoplankton species, M. Stravs, F. Pomati, J. Hollender, Environ. Sci.: Processes Impacts, 2017, 19, 822–832.
Keeping afloat in modern water analysis, A. Sage, The Analytical Scientist, 2017, 22 - 23.
Nontargeted homologue series extraction from hyphenated high resolution mass spectrometry data, M. Loos, H. Singer, J Cheminform 2017, 9:12.
Open-source workflow for smart biotransformation product elucidation using LC-HRMS data, M. Stravs, J. Hollender, CEST 2017, 01416.
Unravelling contaminants in the anthropocene using statistical analysis of liquid chromatography−high-resolution mass spectrometry nontarget screening data recorded in lake sediments, A. Chiaia-Hernandez, B. Guenthardt, M. Frey, J. Hollender, Environ. Sci. Technol. 2017, 51, 12547-12556.
Effect-directed analysis supporting monitoring of aquatic environments — an in-depth overview, W. Brack et al., Science of the Total Environment 544, 2016, 1073–1118.
Rapid screening for exposure to “non-target” pharmaceuticals from wastewater effluents by combining HRMS-based suspect screening and exposure modeling, H. Singer, A. Wössner, C. McArdell, K. Fenner, Environ. Sci. Technol. 2016, 50, 6698−6707.
Vom Unfall zur praeventiven Ueberwachung, J. Mazacek, S. Ruppe, D. Griesshaber, I. Langlois, R. Dolf, H. Singer, J. Leve, A. Hofacker, Aqua & Gas 11, 2016, 66-75.
Prioritizing unknown transformation products from biologically treated wastewater using high-resolution mass spectrometry, multivariate statistics, and metabolic logic, J. Schollee, E. Schymanski, S. Avak, M. Loos, J. Hollender, Anal. Chem. 2015, 87, 12121−12129.
Target screening of chemicals of concern in recycled water, F. Busetti, M. Ruff, K. Linge, Environ. Sci.: Water Res. Technol., 2015, 1, 659.
Targeted and non-targeted liquid chromatography-mass spectrometric workflows for identification of transformation products of emerging pollutants in the aquatic environment, A. Bletsou, J. Jeon, J. Hollender, E. Archontaki, N. Thomaidis, Trends in Analytical Chemistry 66 2015, 32–44.
20 Jahre Rheinueberwachung - Erfolge und analytische Neuausrichtung in Weil am Rhein, M. Ruff, H. Singer, S. Ruppe, J. Mazacek, R. Dolf, C. Leu., Aqua & Gas No 5, 2013, 16 - 25.
Advances in liquid chromatography–high-resolution mass spectrometry for quantitative and qualitative environmental analysis, J. Aceña, S. Stampachiacchiere, S. Pérez, D. Barceló, Anal Bioanal Chem, 2015, 407:6289–6299.
Exploring the behaviour of emerging contaminants in the water cycle using the capabilities of high resolution mass spectrometry, J. Hollender, M. Bourgina, K. Fenner, P. Longrée, C. McArdella, C. Moschet, M. Ruff, E. Schymanskia, H. Singer, Chimia 68, 2014, 793–798.
Investigation of pharmaceuticals and illicit drugs in waters by liquid chromatography-high-resolution mass spectrometry, F. Hernández, M. Ibáñez, R. Bade, L. Bijlsma, J.V. Sancho, Trends in Analytical Chemistry 63, 2014, 140–157.
Iodinated contrast media electro-degradation: Process performance and degradation pathways, G. Moro, C. Pastore, C. Iaconi, G. Mascolo, Science of the Total Environment 506–507, 2015, 631–643.
Plasma lipidomic profiling method based on ultrasound extraction and liquid chromatography mass spectrometry, C. Pizarro, I. Arenzana-Ramila, N. Perez-del-Notario, P. Perez-Matute, J. Gonzalez-Saiz, Anal. Chem. 2013, 85, 12085−12092.
Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry, E. Schymanski, H. Singer, P. Longree, M. Loos, M. Ruff, M. Stravs, C. Vidal, J. Hollender, Environ. Sci. Technol. 2014, 48, 1811−1818.
Screening for pharmaceutical transformation products formed in river sediment by combining ultrahigh performance liquid chromatography/high resolution mass spectrometry with a rapid data-processing method, Z. Li, M. Maier, M. Radke, Analytica Chimica Acta 810 2014, 61–70.
State-of-the-art of screening methods for the rapid identification of chemicals in drinking water, M. Llorca, S. Rodríguez-Mozaz, Report EUR 26155 EN, European Comission 2013.
Suspect and nontarget screening approaches to identify organic contaminant records in lake sediments, A. Chiaia-Hernandez, E. Schymanski, P. Kumar, H. Singer, J. Hollender, Anal Bioanal Chem, 2014, 406:7323–7335.