 v4.3
enviMass
v4.3
enviMass
 v4.3
enviMass
v4.3
enviMass
→ Back to topic overview.
msconvert tool. The latter tool can be downloaded from
→ ProteoWizard and then installed.
Otherwise, contact your LC-MS instrument vendor.
→Back to topic overview.
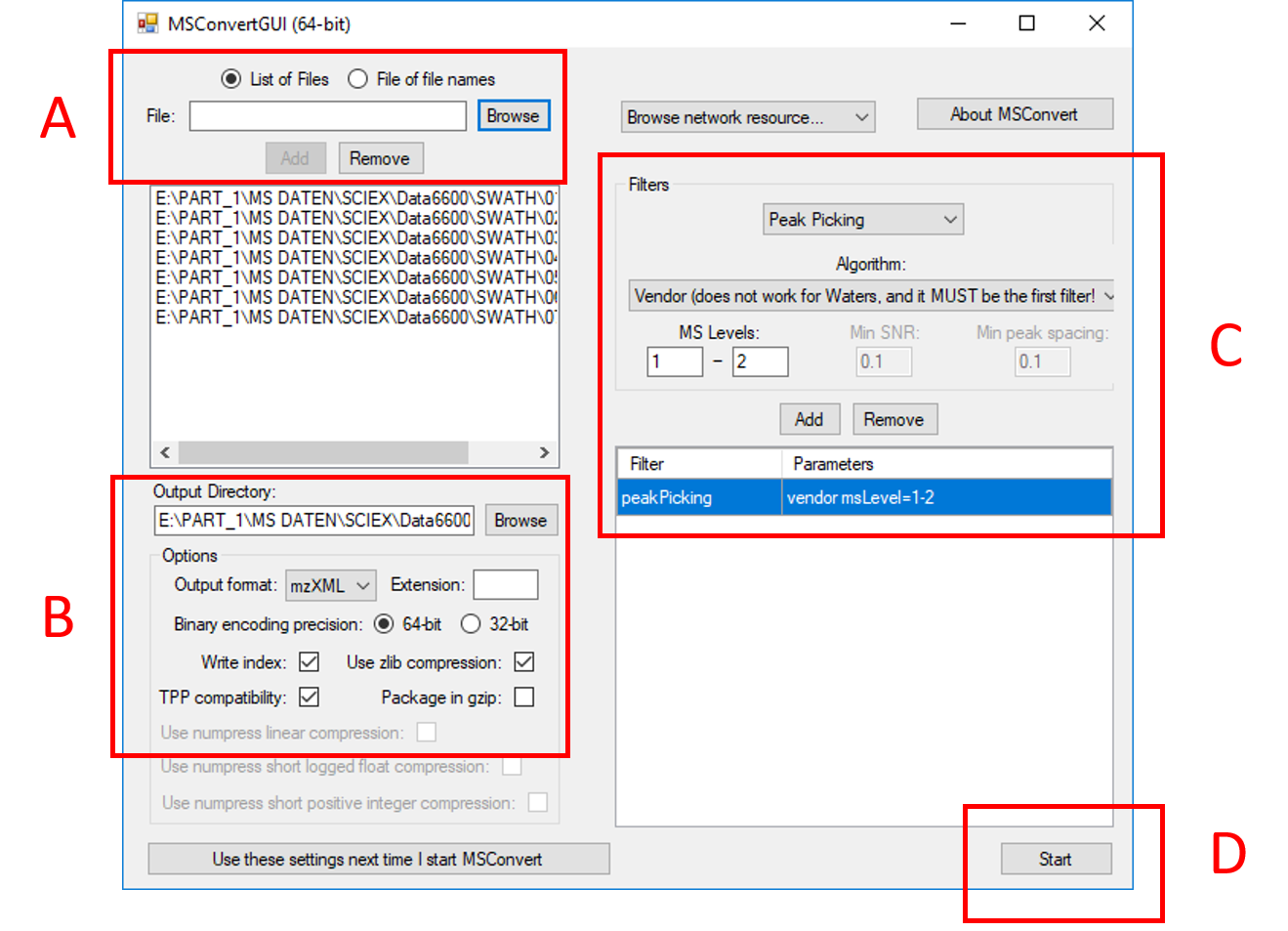 Having installed
Having installed MSConvert, you may use it via command-line or through its GUI, as described here. Four settings in the msconvert GUI are to be included
for file conversion to the centroided .mzXML format, cp. points (A) to (D) in the GUI screenshot:
Add button to include them into your file selection.peakPicking (not to be confused with the enviMass peak picking from the extracted chromatograms) and can be selected from the Filters drop-down selection.
Add button to include this filter during file conversion.
MSConvert tool installed. To do so, users must register the path to msconvert.exe in each of your
projects, as described in the → loading files, File formats section.
→Back to topic overview.
Waters MassLynx
software, run a (lock-mass correction and) file centroidization with the automatic peak detection option (here, peak detection refers to centroidization)._FUNC003 files .DAT, .IDX, .STS) in each resulting data folder. It is not yet clear if this step is always compulsory.ProteoWizard MSConvert
(with Filters → Threshold Peak Filter → Threshold type: Count, Orientation: Most intense, Value: 500(?) → Add; compare also to the steps A, B and D (not C!) described above for Sciex, Thermo or Agilent formats),
massWOLF
(options --mzXML --MSe) or
X2XML
(options unknown).
Workflow tab, press the Apply
and then the Calculate button), move to the Data viewer tab once the calculation has finished and select a file to view its centroid data points plus any peaks picked from them.
Namely, after (a) having enabled the visualization of raw data centroids and (b) having zoomed far enough into an RT and/or m/z range to see the individual centroid points (gray and red dots), is there any strong mass discretization over RT visible between
centoids adjacent in mass? There shouldn`t be any. Are any of the centroid data points highlighted in red, indicating that peaks of reasonable chromatographic shape have been picked? There should be at least some ...
→Back to topic overview.
msconvert for subsequent usage in enviMass has so far only been tested for files from Sciex, Thermo and some Agilent instruments.